Copyright 2021-2023 Lawrence Livermore National Security, LLC and other MuyGPyS Project Developers. See the top-level COPYRIGHT file for details.
SPDX-License-Identifier: MIT
Loss Function Tutorial
This notebook illustrates the loss functions available in the MuyGPyS library. These functions are used to formulate the objective function to be optimized while fitting hyperparameters, and so have a large effect on the outcome of workflows. We will describe each of these loss functions and plot their behaviors to help the user to select the right loss for their problem.
We assume throughout a vector of targets \(y\), a prediction (posterior mean) vector \(\mu\), and a posterior variance vector \(\sigma\) for a training batch \(B\) with \(b\) elements.
[2]:
import matplotlib.pyplot as plt
import numpy as np
from matplotlib.colors import SymLogNorm
from MuyGPyS.optimize.loss import mse_fn, cross_entropy_fn, lool_fn, lool_fn_unscaled, pseudo_huber_fn, looph_fn
from MuyGPyS._src.optimize.loss.numpy import _looph_fn_unscaled
[3]:
mmax = 3.0
point_count = 50
ys = np.zeros(point_count)
mus = np.linspace(-mmax, mmax, point_count)
smax = 3.0
smin = 1e-1
sigma_count = 50
sigmas = np.linspace(smin, smax, sigma_count)
Variance-free Loss Functions
MuyGPyS features several loss functions that depend only upon \(y\) and \(\mu\) of your training batch. These loss functions are situationally useful, although they leave the fitting of variance parameters entirely up to the separate, analytic sigma_sq optimization function and might not be sensitive to certain variance parameters. As they do not require evaluating \(\sigma\) or optimizing the variance scaling parameter, these loss functions are generally more efficient to
use in practice.
Mean Squared Error (mse_fn)
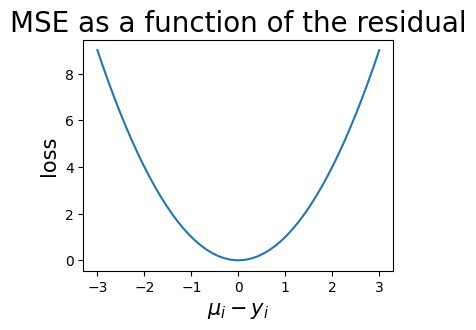
The mean squared error (MSE) is a classic loss that computes
The string used to indicate MSE loss in optimization functions is "mse". The following plot illustrates the MSE as a function of the residual.
[4]:
fig, ax = plt.subplots(1, 1, figsize=(4,3))
ax.set_title("MSE as a function of the residual", fontsize=20)
ax.set_ylabel("loss", fontsize=15)
ax.set_xlabel("$\mu_i - y_i$", fontsize=15)
mses = [mse_fn(ys[i].reshape(1, 1), mus[i].reshape(1, 1)) for i in range(point_count)]
ax.plot(mus, mses)
plt.show()
Cross Entropy Loss (cross_entropy_fn)
The cross entropy loss is a classic classification loss often used in the fitting of neural networks. Optimization functions recognize "cross_entropy" or "log" as arguments to loss_method.
⚠️ This section is under construction. ⚠️
Pseudo-Huber Loss (pseudo_huber_fn)
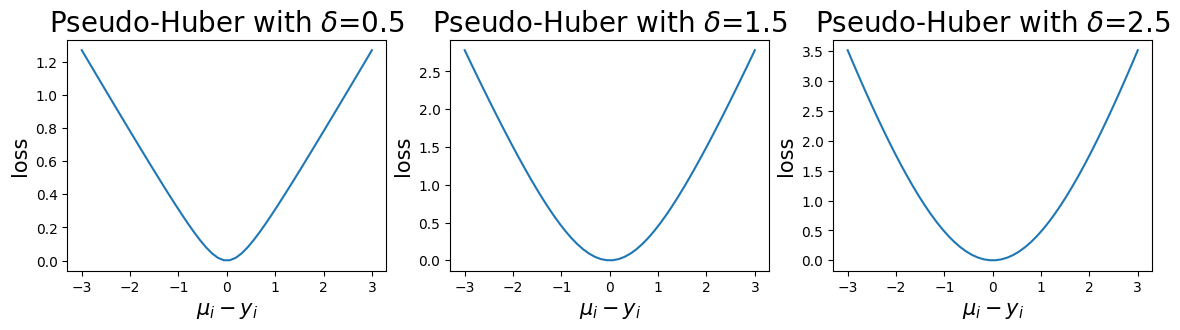
The pseudo-Huber loss is a smooth approximation to the Huber loss, which is approximately quadratic for small residuals and approximately linear for large residuals. This means that the pseudo-Huber loss is less sensitive to large outliers, which might otherwise force the optimizer to overcompensate in undesirable ways. The pseudo-Huber loss computes
where \(\delta\) is a parameter that indicates the scale of the boundary between the quadratic and linear parts of the function. The pseudo_huber_fn accepts this parameter as the boundary_scale keyword argument. Note that the scale of \(\delta\) depends on the units of \(y\) and \(mu\). The following plots show the behavior of the pseudo-Huber loss for a few values of \(\delta\).
[5]:
boundary_scales = [0.5, 1.5, 2.5]
phs = np.array([
[pseudo_huber_fn(ys[i].reshape(1, 1), mus[i].reshape(1, 1), boundary_scale=bs) for i in range(point_count)]
for bs in boundary_scales
])
fig, axes = plt.subplots(1, 3, figsize=(14, 3))
for i, ax in enumerate(axes):
ax.set_title(f"Pseudo-Huber with $\delta$={boundary_scales[i]}", fontsize=20)
ax.set_ylabel("loss", fontsize=15)
ax.set_xlabel("$\mu_i - y_i$", fontsize=15)
ax.plot(mus, phs[i, :])
plt.show()
Variance-Sensitive Loss Functions
MuyGPyS also includes loss functions that explicitly depend upon the posterior variances (\(\sigma\)). These loss functions penalize large variances, and so tend to be more sensitive to variance parameters. This comes at increasing the cost of the linear algebra involved in each evaluation of the objective function by a constant factor. This causes an overall increase in compute time per optimization loop, but that is often worth the trade for sensitivity in practice.
\(\sigma\) involves multiplying the unscaled MuyGPS variance by the sigma_sq variance scaling parameter, which at present must by optimized during each evaluation of the objective function.
Leave-One-Out Loss (lool_fn)
The leave-one-out-loss or lool scales and regularizes the MSE to make the loss more sensitive to parameters that primarily act on the variance. lool computes
The next plot illustrates the loss as a function of both the residual and of \(\sigma\).
[6]:
lools = np.array([
[
lool_fn_unscaled(
ys[i].reshape(1, 1),
mus[i].reshape(1, 1),
sigmas[sigma_count - 1 - j]
)
for i in range(point_count)
]
for j in range(sigma_count)
])
[7]:
fig, axes = plt.subplots(1, 4, figsize=(19,4))
axes[0].set_title("lool", fontsize=20)
axes[0].set_ylabel("$\sigma_i$", fontsize=15)
axes[0].set_xlabel("$\mu_i - y_i$", fontsize=15)
im = axes[0].imshow(
lools, extent=[-mmax, mmax, smin, smax], norm=SymLogNorm(0.5), cmap="coolwarm"
)
fig.colorbar(im, ax=axes[0])
axes[1].set_title("lool, $\sigma_i=0.5$", fontsize=20)
axes[1].set_ylabel("lool", fontsize=15)
axes[1].set_xlabel("$\mu_i - y_i$", fontsize=15)
axes[1].plot(mus, lools[7, :])
axes[2].set_title("lool, $\mid \mu_i - y_i \mid=1.0$", fontsize=20)
axes[2].set_ylabel("lool", fontsize=15)
axes[2].set_xlabel("$\sigma_i$", fontsize=15)
axes[2].plot(sigmas, np.flip(lools[:, 33]))
axes[3].set_title("lool, $\mid \mu_i - y_i \mid=0.0$", fontsize=20)
axes[3].set_ylabel("lool", fontsize=15)
axes[3].set_xlabel("$\sigma_i$", fontsize=15)
axes[3].plot(sigmas, np.flip(lools[:, 24]))
plt.tight_layout()
plt.show()
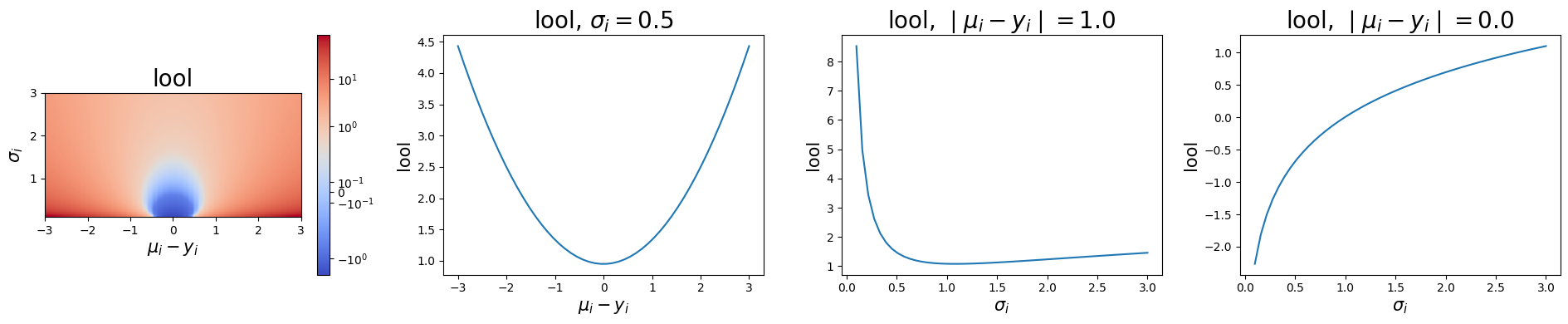
Notice that the cross-section of the lool surface for a fixed \(\sigma\) is quadratic, while the cross section of the lool surface for a fixed residual is logarithmic. For small enough residuals, this curve inverts and assumes negative values for small \(\sigma\).
Leave-One-Out Pseudo-Huber (looph_fn)
The leave-one-out pseudo-Huber loss (looph) is similar in nature to the lool, but is applied to the pseudo-Huber loss instead of MSE. looph computes
where again \(\delta\) is the boundary scale. The next plots illustrate the looph as a function of the residual, \(\sigma\), and \(\delta\).
[8]:
loophs = np.array([
[
[
_looph_fn_unscaled(
ys[i].reshape(1, 1),
mus[i].reshape(1, 1),
sigmas[sigma_count - 1 - j],
boundary_scale=bs
)
for i in range(point_count)
]
for j in range(sigma_count)
]
for bs in boundary_scales
])
[9]:
fig, axes = plt.subplots(3, 4, figsize=(19,14))
for i, bs in enumerate(boundary_scales):
axes[i, 0].set_title(f"looph, $\delta={bs}$", fontsize=20)
axes[i, 0].set_ylabel("$\sigma_i$", fontsize=15)
axes[i, 0].set_xlabel("$\mu_i - y_i$", fontsize=15)
im = axes[i, 0].imshow(
lools, extent=[-mmax, mmax, smin, smax], norm=SymLogNorm(0.5), cmap="coolwarm"
)
fig.colorbar(im, ax=axes[i, 0])
axes[i, 1].set_title(f"looph, $\delta={bs}$, $\sigma_i=0.5$", fontsize=20)
axes[i, 1].set_ylabel("looph", fontsize=15)
axes[i, 1].set_xlabel("$\mu_i - y_i$", fontsize=15)
axes[i, 1].plot(mus, loophs[i, 7, :])
axes[i, 2].set_title(f"looph, $\delta={bs}$, $\mid \mu_i - y_i \mid=1.0$", fontsize=20)
axes[i, 2].set_ylabel("looph", fontsize=15)
axes[i, 2].set_xlabel("$\sigma_i$", fontsize=15)
axes[i, 2].plot(sigmas, np.flip(loophs[i, :, 33]))
axes[i, 3].set_title(f"looph, $\delta={bs}$, $\mid \mu_i - y_i \mid=0.0$", fontsize=20)
axes[i, 3].set_ylabel("looph", fontsize=15)
axes[i, 3].set_xlabel("$\sigma_i$", fontsize=15)
axes[i, 3].plot(sigmas, np.flip(loophs[i, :, 24]))
plt.tight_layout()
plt.show()
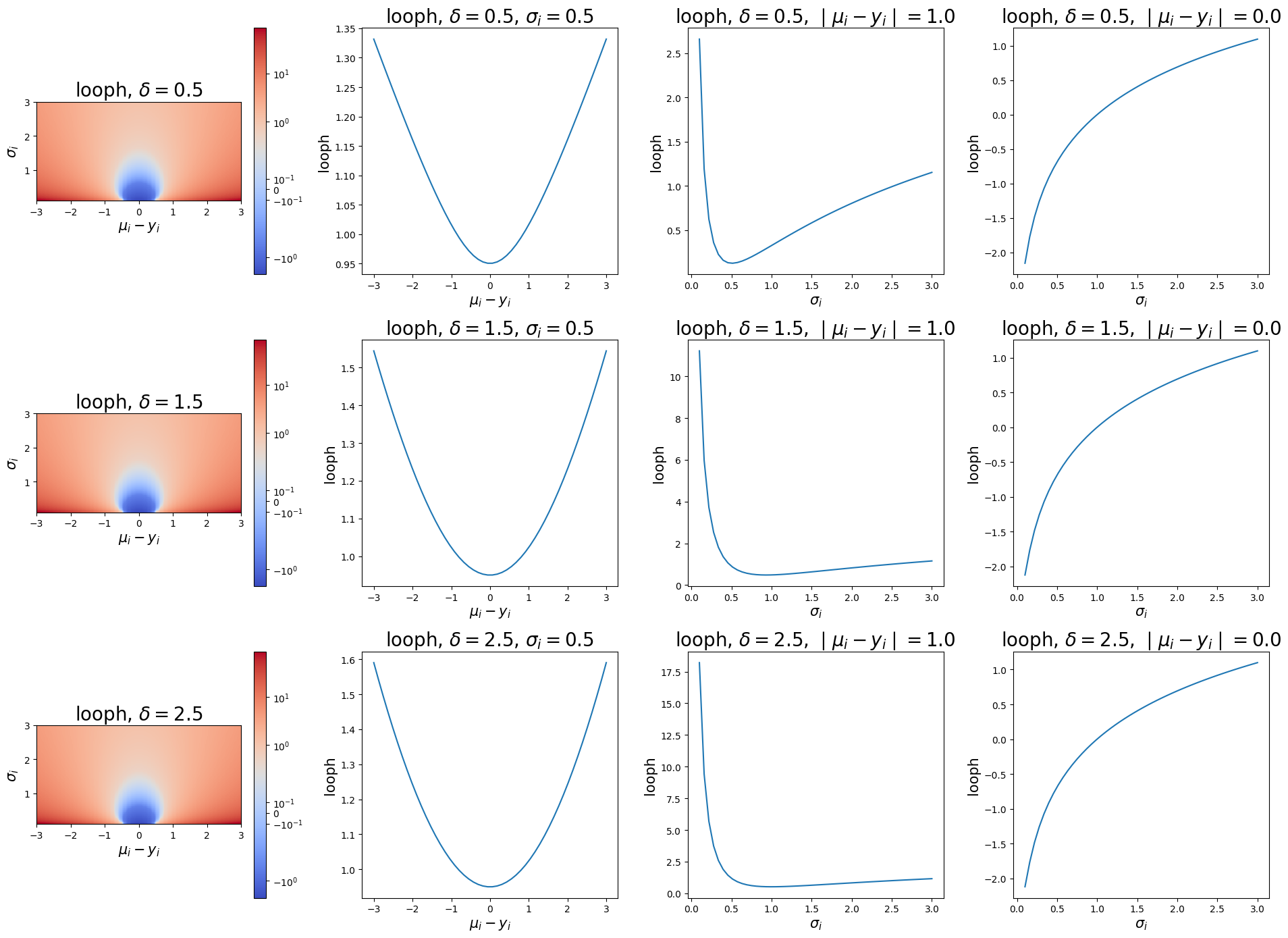
These plots show us that the looph function can exhibit a more exaggerated upward slope where the residual is in the linear component of the pseudo-Huber curve but is not so large that it still outweighs the variance component of the loss. Note that in practice that both pseudo Huber loss functions may require more training iterations to converge than their alternatives.